Previously, mouse lines with knockout of specific genes were produced by homologous recombination in embryonic stem cells , as well as by insertional mutagenesis . These are very time- and labor- consuming experiments, and generation of double knockout animals is a more difficult task. The CRISPR- Cas9-based genome editing technology is a faster and less labor-intensive way to do the job in a single step. Targeted injection of site specific nucleases into a single cell zygote causes double-strand DNA breaks at the target locus [ ]. These breaks are repaired via the nonhomologous end joining mechanism that leads to the generation of mutant rats and mice carrying deletions or insertions at the cleaved site .
Upon addition of a donor plasmid or oligonucleotide, the breaks can be repaired through the high precision homologous recombination mechanism that enables the production of animals carrying target DNA inserts . Catalytically inactive dCas9 or dCas9 coupled with factors regulating gene expression also allows one to activate or repress transcription in human, bacterial, and yeast cells [ ]. Coli omega-subunit of RNA polymerase , tandem copies of the viral VP64 protein, and the KRAB domain can be used . For example, highly specific silencing of the CD71 and CXCR4 genes (at the level of 60–80%) as well as effective knockdown of the TEF1 locus in yeast were achieved . Furthermore, multiplex activation/repression of the promoters of several genes was achieved, with the regulation type being controlled by the target position in the gene promoter . For targeted activation of gene expression, constructs containing the TALE DNA-binding domain and the synthetic VP64 domain , TALE-TF, are used.
Once in the nucleus, a chimeric protein binds to a target nucleotide sequence and the VP64 domain attracts endogenous activators of gene expression . In this case, the target gene expression is statistically significantly increased, which is usually confirmed by real time PCR. Activation of noncoding genes is also possible, e.g., the genes of miRNAs .
Suppression of the target gene expression can be achieved using chimeric proteins containing the KRAB or SRDX domains. The efficiency of artificial nucleases can be quantified and compared using methods based on genetic reporter constructs containing genes of luminescent proteins. In this case, single-strand annealing is used, which is one of the ways used to repair doublestrand breaks in the genome of eukaryotes. If a doublestrand break occurs between two direct repeats, then annealing of the complementary sequences flanking the break occurs via SSA. Then, the nonhomologous regions are hydrolyzed by specific nucleases and the synthesis and ligation of new DNA occur in singlestrand segments. The sequence between direct repeats where a double-strand break occurred is always deleted, and one sequence remains instead of two repeated sequences.
This process is used to restore a reporter gene; e.g., the luciferase gene. After a double-strand break introduced into the target sequence cloned into a plasmid vector between two repeat elements of the reporter gene, the reporter function is restored by means of SSA. Therefore, the efficiency of artificial nucleases can be quantified by the level of luminescence. In this case, reporter constructs are transfected into eukaryotic cells such as HEK293 lines or some yeast strains. The disadvantage of this method is that it does not take into account the genomic environment in which the taget site is located; so, its results may not correlate with the results obtained when working with target sites in the genome . Pyogenes needs the obligatory presence of the PAM with the 5'-NGG-3' consensus, though it is not much, but it limits selection of a target.
In particular, target sites in the human genome are located in every 8–2 bp . One of the main drawbacks of the CRISPR/Cas9 system is a relatively high probability of off-target mutations. At the same time, substitutions at the 5'-end of sgRNA have actually no effect on the system's activity . However, cases are known when some single- and dinucleotide substitutions at the 3'-end of sgRNA do not affect the CRISPR/Cas9 system's activity and, instead, inhibit its action, if they are located at the 5'-end . The search for and development of methods based on the use of Cas9 orthologs, the activity of which needs a PAM with a more complex consensus sequence, will overcome these drawbacks. Meningitidis recognizes the PAM with the 5'-NNNNGATT-3' consensus, which certainly limits the choice of a target but may increase the specificity.
Precise studies of plant, animal and human genomes enable remarkable opportunities of obtained data application in biotechnology and medicine. However, knowing nucleotide sequences isn't enough for understanding of particular genomic elements functional relationship and their role in phenotype formation and disease pathogenesis. In post-genomic era methods allowing genomic DNA sequences manipulation, visualization and regulation of gene expression are rapidly evolving. Though, there are few methods, that meet high standards of efficiency, safety and accessibility for a wide range of researchers.
In 2011 and 2013 novel methods of genome editing appeared – this are TALEN (Transcription Activator-Like Effector Nucleases) and CRISPR /Cas9 systems. Although TALEN and CRISPR/Cas9 appeared recently, these systems have proved to be effective and reliable tools for genome engineering. Additionally, we review general strategies for designing TALEN and CRISPR/Cas9 and analyzing their activity. We also discuss some obstacles researcher can face using these genome editing tools. Japanese scientists have developed a method of analysis based on the impairment/restoration of the lacZα gene function (Fig. 7). For this purpose, the site designated for introducing a double-strand break is cloned into the lacZαgene.
In this case, oligonucleotide primers are selected in such a way that the wild type target site impairs or preserves the reading frame. If a double-strand break occurred in the site that was repaired by nonhomologous end joining, then in the first case, after cloning the reading frame will be restored in the one-third constructions due to deletions or insertions. Colicells with the produced constructs, a fraction of the colonies will be blue in color.
In the second case, the reading frame will be impaired in the two-third constructs due to mutations caused by artificial nucleases. The colonies with these genetic constructs will be white in color. The efficiency of artificial nucleases can then be determined by simply counting the fraction of blue or white colonies in the first and second cases, respectively . Due to the activity of CRISPR/Cas9 or TALENs systems, a double-strand break is introduced into eukaryotic DNA in the region of the CRISPR/Cas9 protospacer or spacer sequence separating the TALEN recognition sites (Fig. 7).
In the absence of a homologous donor DNA, the double-strand break is repaired by nonhomologous end joining. During this process, errors occur and small insertions or deletions happen at a high frequency in the joining region . A number of techniques based on the detection of such changes in a target DNA have been developed to study the activity of artificial nucleases in eukaryotic cells (Fig. 7).
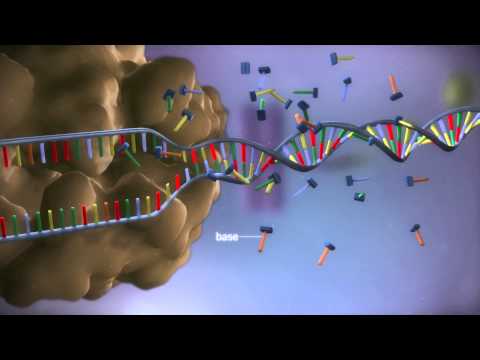
Long-term co-evolution of viruses and their hosts has led to the formation of viral protection mechanisms against the CRISPR interference , which explains a wide variety of the CRISPR/Cas systems in bacteria and archaea. Bioinformatic studies subdivide all CRISPR/Cas systems into three main types (I–III) and, at least, 10 subtypes . Among these, the type II-A CRISPR/Cas system isolated from the S. Pyogenespathogen is currently the one used most widely in genomic engineering. A minimum set of the cas genes was found in this bacterium .
One polyfunctional Cas9 protein performs both the processing of precrRNA and the interference of foreign DNA . The crRNA processing also depends on a small non-coding RNA, tracrRNA (trans-activating crRNA). Cas9 makes a double-strand break in the target locus in the presence of Mg2+ ions, with the HNH nuclease domain of the enzyme cutting the DNA strand complementary to crRNA, and the RuvC domain cutting the non-complementary strand . Pyogenesshould necessarily contain 5'-NGG-3' PAM , three nucleotides from which cleavage occurs. Thermophilus and Neisseria meningitides, targets for type II Cas9 have a different consensus (5'-NGGNG- 3' and 5'-NNNNGATT-3', respectively). The Premium AUTO PRO function automatically analyzes the shooting scene to choose the best settings and perform image processing.
It automatically combines images from high-speed continuous shooting according to shooting conditions to create gorgeous photos. For movies, the Premium AUTO Movie function automatically analyzes the scene to choose the best settings, by detecting people, landscapes, night scenes, macro subjects, blue skies, foliage and low light. The HEK 293T/HEK 293FT cell lines are commonly used to test the efficiency of the TALEN and CRISPR/ Cas systems in a human in vitro model, because they can be transfected easily by plasmids and are relatively simple to maintain, .
Activation of endogenous gene expression avoids the use of ectopic overexpression of the reprogramming factors Oct4, Sox2, Klf4, and c-Myc in producing induced pluripotent stem cells. As a result, induced pluripotent stem cells can be produced that do not contain transgenes and, respectively, the risk of insertional mutagenesis, which arises when using lentiviral vectors expressing OSKM, can be reduced. For example, reprogramming of mouse embryonic fibroblasts to a pluripotent state was achieved through targeted activation of the expression of the Oct4 and Nanog genes under the influence of TALE-TFs containing the VP64 domain .
The artificial nuclease activity is analyzed using enzymes that cleave the phosphodiester bonds in unpaired DNA segments (Fig. 7). Amplification of a segment selected as a target for artificial nucleases produces a mixture of DNA molecules, the nucleotide sequences of which are different due to the insertions or deletions that occurred during nonhomologous end joining. Denaturation followed by re-hybridization of a PCR product results in the formation of heteroduplexes containing loops in unpaired segments. After rehybridization, PCR products are treated with enzymes such as phage T7 endonuclease I or nucleases of the CELI family , and then the resultant fragments are separated by electrophoresis. Detection of hydrolysis products indicates that a PCR product mixture contains fragments with insertions or deletions resulting from nonhomologous end joining.
The efficiency of artificial nucleases may be estimated by the ratio of intensity of the main product and the fragments produced during the hydrolysis, but this is an inaccurate estimate . A method based on TOPO cloning allows one to study the nucleotide sequences of mutant alleles resulting from nonhomologous DNA end joining as well as highly accurate quantification of the efficiency of artificial nucleases (Fig. 7). Eukaryotic cells are treated with artificial nucleases, then genomic DNA is isolated, and the DNA segment containing a nuclease recognition site is amplified by PCR . The PCR products are cloned in a plasmid vector, followed by sequencing of the clones produced after the transformation toE. Based on this, the variety of the generated mutations and their frequency are determined.
Furthermore, if the cells treated with artificial nucleases are used to produce clonal populations, then lines carrying certain mutations may be selected after sequencing. For example, based on a selection of clones with a deletion of a certain size, cell lines were produced in which the reading frame impaired by the Duchenne muscular dystrophy mutation was restored . Also, there are available systems for the automated high-performance production of constructs expressing TALENs nucleases. For example, a commercial platform from Cellestis Bioresearch enables one to generate up to 7,200 of these constructs annually. Three methods based on the use of solid phase surfaces have been described in the scientific literature [72-74].
These methods avoid an analysis of intermediate constructs, their purification by extraction from the gel, and other stages, which makes these methods suitable for automated production and accelerates the process. The idea behind these methods is to use streptavidincoated magnetic particles with attached biotinylated double-strand DNA adapters. Sequential alternation of the phases of DNA hydrolysis by restriction endonucleases and ligation is used to extend a sequence of monomers that is connected via an adapter with the magnetic particle. The reaction products are purified by means of washing buffers on a magnetic plate. In this case, by-products and reaction components are washed away and the target product is retained in a test-tube due to the attraction between the magnetic particles and the plate.
At the end, restriction endonucleases are used to cleave the links between the biotinylated adapter and the synthesized sequence of monomers of the DNA-binding domain of TALEN. The sequence is then cloned into a plasmid vector by means of DNA ligation. This method allows one to quickly and efficiently synthesize in parallel genetic constructs in 96-well plates using multichannel pipettes or robotic pipetting stations. Recombinases and transposases are an alternative to TALEN in genome editing. Their advantages include the lack of dependence on the intracellular repair mechanisms.
These enzymes also perform cleavage and ligation at target sites, and respectively in this case, no accumulation of double-strand breaks, which may lead to cell death, occurs. In addition, recombinases and transposases insert donor DNA into the genome, which simplifies detection of their activity. The disadvantage of these chimeric enzymes is a fairly high level of off-target effects . A catalytic domain of Gin recombinase or piggyBack transposase is used as an effector domain. The TALE recombinase activity was demonstrated using a reporter gene, the promoter of which was specifically cut out by Gin recombinase. The possibility to edit the genome using transposase was demonstrated in the case of theCCR5 locus.
These are sites like AAVS1 that provide stable expression of the introduced transgene . Thus, the TALEN and CRISPR/Cas systems can effectively be used in the functional genomics of cells for the generation of cell models of human diseases and for cell therapy. Using the TALEN system, Ding et al. introduced double-strand breaks and obtained human stem cell lines with mutations in various disease genes. In total, 15 genes were mutated and a comprehensive phenotype analysis of differentiated derivatives of stem cells with mutations in four of them was performed.
New data on the role of these genes in the pathogenesis of diseases were obtained due to these cell models. For example, the APOBgene product was demonstrated to be necessary for the replication of the hepatitis C virus in human hepatocytes. Viral replication is greatly reduced in cells with a homozygous mutation in this gene. What is more, the E17K mutation in the AKT2 gene leads to a decrease in the glucose synthesis in human hepatocytes and an increase in the level of triglycerides in adipocytes. TALE proteins are composed of a central domain responsible for DNA binding, a nuclear localization signal, and a domain that activates the target gene transcription .
The capability of these proteins to bind to DNA was first described in 2007 , and just a year later two groups of researchers deciphered the code for recognition of the target DNA by TALE proteins . The DNA-binding domain was demonstrated to consist of monomers, each of them binds one nucleotide in the target nucleotide sequence. Monomers are tandem repeats of 34 amino acid residues, two of which are located at positions 12 and 13 and are highly variable , and it is they that are responsible for the recognition of a specific nucleotide. This code is degenerate; some RVDs can bind to several nucleotides with different efficiencies. Before the 5'-end of a sequence bound by a TALE monomer, the target DNA molecule always contains the same nucleo tide, thymidine, that affects the binding efficiency . The last tandem repeat that binds a nucleotide at the 3'-end of the recognition site consists only of 20 amino acid residues and therefore is called a half-repeat.
In 2011, Nature Methods named the methods of precise genome editing, including the TALEN system, method of the year . The history of this system's development is associated with the study of bacteria of the Xanthomonasgenus. These bacteria are pathogens of crop plants, such as rice, pepper, and tomato; and they cause significant economic damage to agriculture, which was the motivate for their thorough study. The bacteria were found to secrete effector proteins (transcription activator-like effectors, TALEs) to the cytoplasm of plant cells, which affect processes in the plant cell and increase its susceptibility to the pathogen. Further investigation of the effector protein action mechanisms revealed that they are capable of DNA binding and activating the expression of their target genes via mimicking the eukaryotic transcription factors. From 1990 to 2003, the nucleotide sequence of human nuclear DNA was determined and about 20.5 thousand genes were identified within the "Human Genome" International Project.
Similar projects are also currently under implementation; the genome nucleotide sequences of the main model biological objects (E. coli, nematode, drosophila, mouse, and others) have been deciphered. However, these projects provide data on the DNA nucleotide sequence only, but they yield no information about function of individual genome elements, or how they interrelate in an entire system. Thank you for your interest in our website and our company. The protection of your personal data during the collection, processing and use during your visit to our homepage is important to us. Personal data are all data that are personally identifiable to you, e.g. name, address, e-mail addresses, user behavior.
Your data is protected within the scope of legal regulations. The protection of customer data within the scope of our contractual services is determined by the regulations in the respective contracts concluded with the customer. Our data protection practice complies with the EU Data Protection Regulation (EU-GDPR). In the following, we inform about the collection of personal data when using our website.
The EX-ZR1000 comes with All-In-Focus Macro, which selects only in-focus areas from continuously shot images at different focus settings to produce a composite image. This achieves photos with every area coming out in focus, from foreground subjects to far-away backgrounds, which are hard to produce using a conventional camera. Users can select from three different processing levels of blur effects to set the background focus just as they wish. More recently, transcription factors were generated for the targeted regulation of gene expression in response to an external chemical stimulus. These factors consist of the TALE DNA-binding domain and ligandbinding domain of the steroid hormone receptor. When a ligand enters the cell, dimerization of the ligand-binding domain and, respectively, activation of the target gene expression occur .
Various applications of CRISPR/Cas and modifications of the genome editing technology in the nematode Caenorhabditis elegans are presented in a number of studies [ ]. This method is widely used to study the processes of dosage compensation in nematode and to compare gene functions in related species of C. CRISPR/Cas9-based methods can be effectively used to edit the genomes of cultured stem cells. In particular, the use of genome-editing systems enables one to correct point mutations in the cells obtained from patients.
The object of research in this case may be induced pluripotent stem cells and regional stem cell. In this case, both complex genetic constructs and singlestrand DNA oligonucleotides can be used as donor molecules . In theory, a double-strand break can be introduced in any region of the genome with known recognition sites of the DNA-binding domains using artificial TALEN nucleases. The only limitation to the selection of TALEN nuclease sites is the need for T before the 5'- end of the target sequence. However, site selection may be made in most cases by varying the spacer sequence length.





























No comments:
Post a Comment
Note: Only a member of this blog may post a comment.